Optimized Hyper-slicing in PyTables with Blosc2 NDim
The recent and long-awaited PyTables 3.9 release carries many goodies, including a particular one which makes us at the PyTables and Blosc teams very excited: optimized HDF5 hyper-slicing that leverages the two-level partitioning schema in Blosc2 NDim. This development was funded by a NumFOCUS grant and the Blosc project.
I (Ivan) carried on with the work that Marta started, with very valuable help from her and Francesc. I was in fact a core PyTables developer quite a few years ago (2004-2008) while working with Francesc and Vicent at Cárabos Coop. V. (see the 20 year anniversary post for more information), and it was an honour and a pleasure to be back at the project. It took me a while to get back to grips with development, but it was a nice surprise to see the code that we worked so hard upon live through the years and get better and more popular. My heartfelt thanks to everybody who made that possible!
Update (2023-11-23): We redid the benchmarks described further below with some fixes and the same versions of Blosc2 HDF5 filter code for both PyTables and h5py. Results are more consistent and easier to interpret now.
Update (2023-12-04): We extended benchmark results with the experimental application of a similar optimization technique to h5py.
Direct chunk access and two-level partitioning
You may remember that the previous version of PyTables (3.8.0) already got support for direct access to Blosc2-compressed chunks (bypassing the HDF5 filter pipeline), with two-level partitioning of chunks into smaller blocks (allowing for fast access to small slices with big chunks). You may want to read Óscar and Francesc's post Blosc2 Meets PyTables to see the great performance gains provided by these techniques.
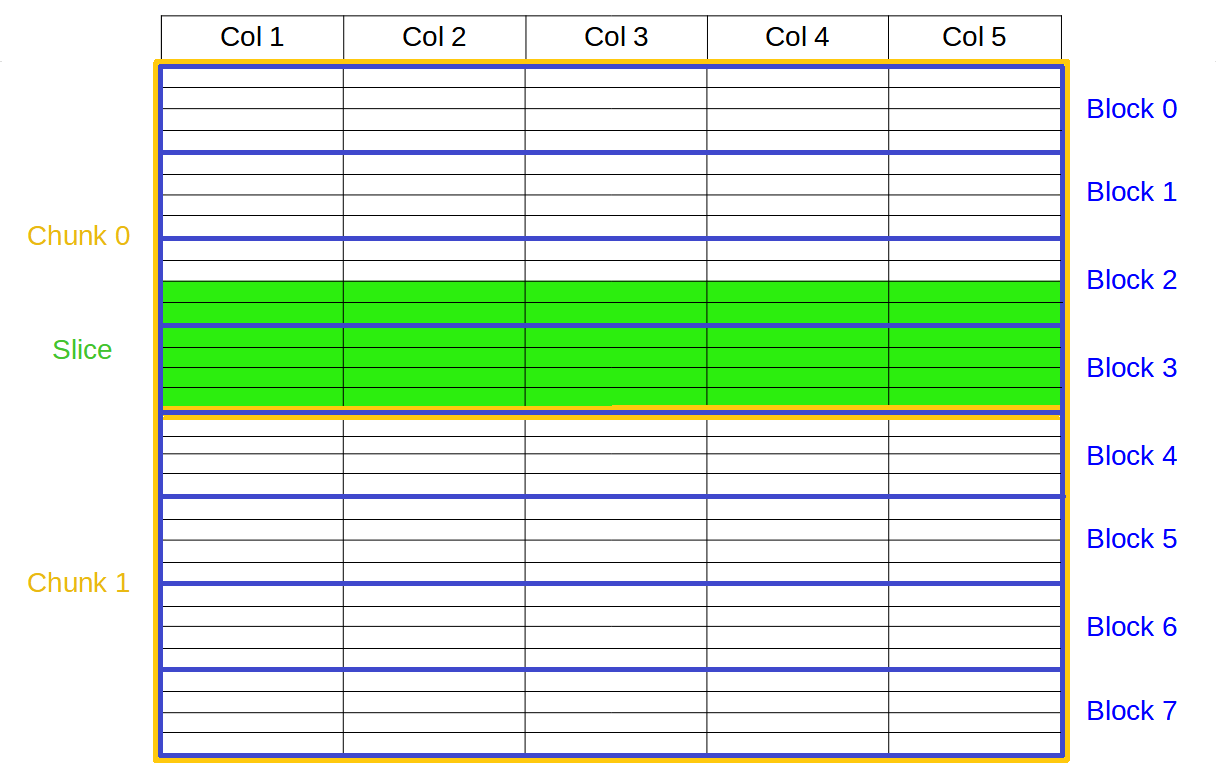
However, these enhancements only applied to tabular datasets, i.e. one-dimensional arrays of a uniform, fixed set of fields (columns) with heterogeneous data types as illustrated above. Multi-dimensional compressed arrays of homogeneous data (another popular feature of PyTables) still used plain chunking going through the HDF5 filter pipeline, and flat chunk compression. Thus, they suffered from the high overhead of the very generic pipeline and the inefficient decompression of whole (maybe big) chunks even for small slices.
Now, you may have also read the post by the Blosc Development Team about Blosc2 NDim (b2nd), first included in C-Blosc 2.7.0. It introduces the generalization of Blosc2's two-level partitioning to multi-dimensional arrays as shown below. This makes arbitrary slicing of such arrays across any dimension very efficient (as better explained in the post about Caterva, the origin of b2nd), when the right chunk and block sizes are chosen.
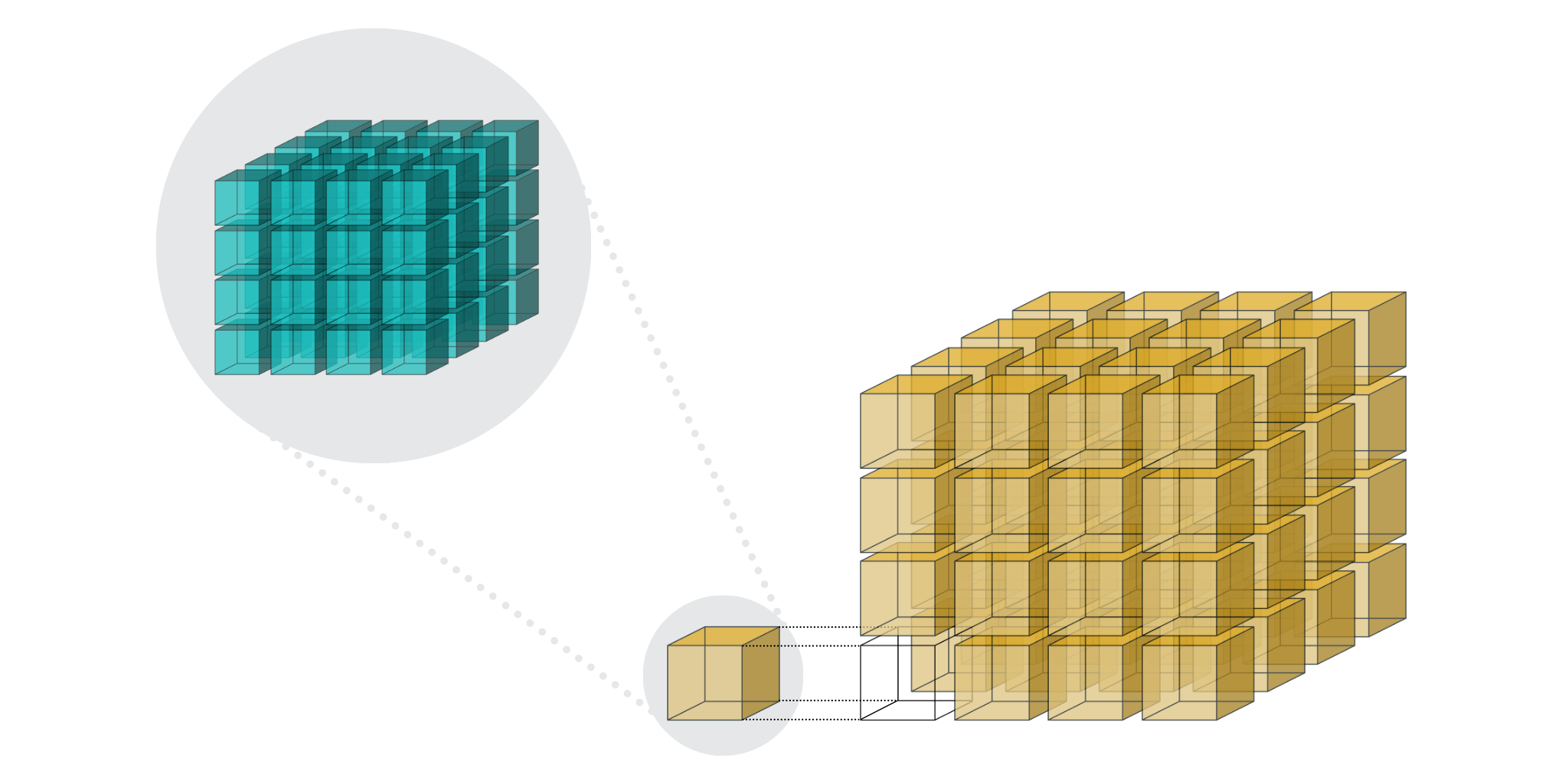
This b2nd support was the missing piece to extend PyTables' chunking and slicing optimizations from tables to uniform arrays.
Choosing adequate chunk and block sizes
Let us try a benchmark very similar to the one in the post introducing Blosc2 NDim, which slices a 50x100x300x250 floating-point array (2.8 GB) along its four dimensions, but this time with 64-bit integers, and using PyTables 3.9 with flat slicing (via the HDF5 filter pipeline), PyTables 3.9 with b2nd slicing (optimized, via direct chunk access implemented in C), h5py 3.10 with flat slicing (via hdf5plugin 4.3's support for Blosc2 in the HDF5 filter pipeline), and h5py with b2nd slicing (via the experimental b2h5py package using direct chunk access implemented in Python through h5py).
According to the aforementioned post, Blosc2 works better when blocks have a size which allows them to fit both compressed and uncompressed in each CPU core’s L2 cache. This of course depends on the data itself and the compression algorithm and parameters chosen. Let us choose LZ4+shuffle since it offers a reasonable speed/size trade-off, and try to find the different compression levels that work well with our CPU (level 8 seems best in our case).
With the benchmark's default 10x25x50x50 chunk shape, and after experimenting with the BLOSC_NTHREADS environment variable to find the number of threads that better exploit Blosc2's parallelism (6 for our CPU), we obtain the results shown below:
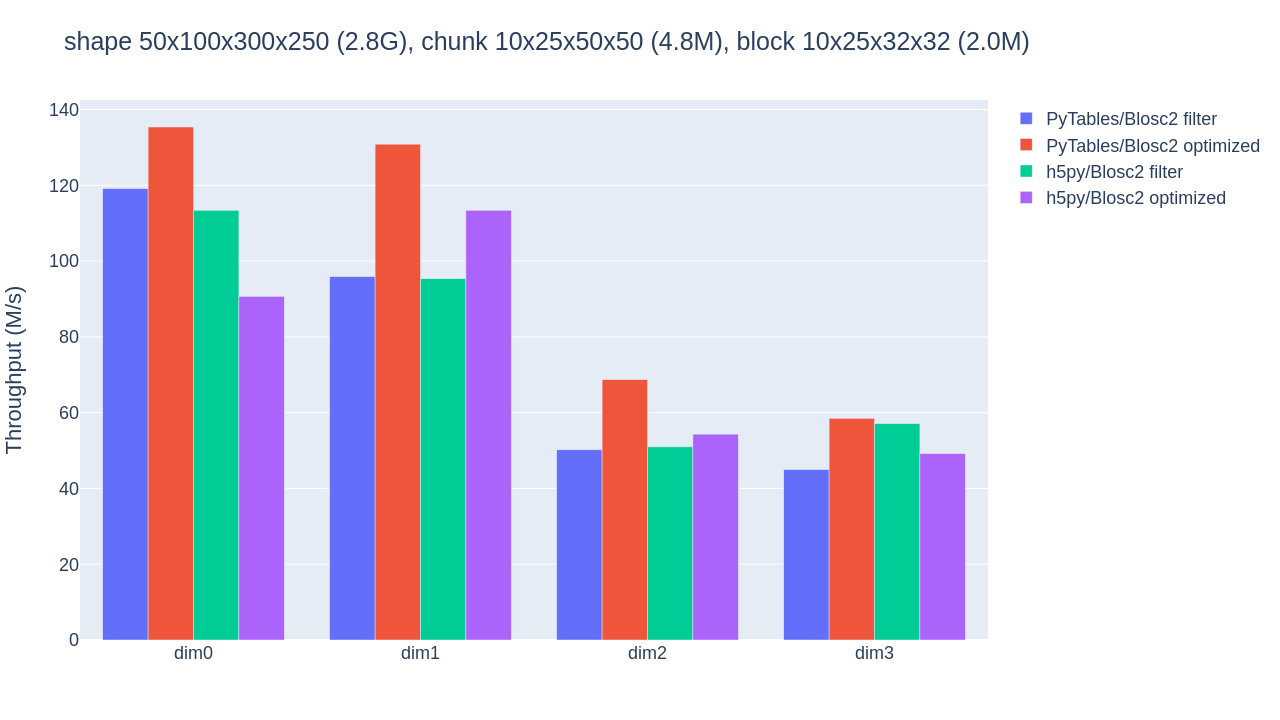
The optimized b2nd slicing of PyTables already provides some speedups (although not that impressive) in the inner dimensions, in comparison with flat slicing based on the HDF5 filter pipeline (which performs similarly for PyTables and h5py). As explained in Blosc2 Meets PyTables, HDF5 handling of chunked datasets favours big chunks that reduce in-memory structures, while Blosc2 can further exploit parallel threads to handle the increased number of blocks. Our CPU's L3 cache is 36MB big, so we may still grow the chunksize to reduce HDF5 overhead (without hurting Blosc2 parallelism).
Let us raise the chunkshape to 10x25x150x100 (28.6MB) and repeat the benchmark (again with 6 Blosc2 threads):
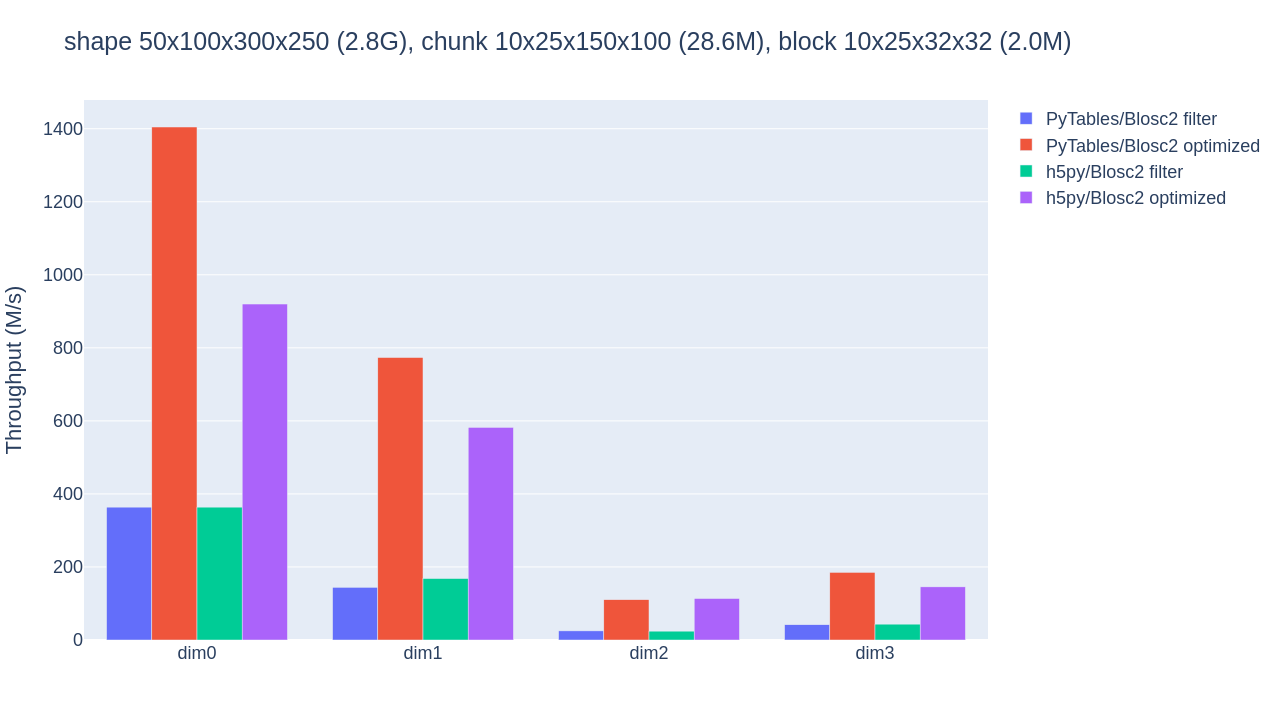
Much better! Choosing a better chunkshape not just provides up to 10x speedup for the PyTables optimized case, it also results in 4x-5x speedups compared to the performance of the HDF5 filter pipeline. The optimizations applied to h5py also yield considerable speedups (for an initial, Python-based implementation).
Conclusions and future work
The benchmarks above show how optimized Blosc2 NDim's two-level partitioning combined with direct HDF5 chunk access can yield considerable performance increases when slicing multi-dimensional Blosc2-compressed arrays under PyTables (and h5py). However, the usual advice holds to invest some effort into fine-tuning some of the parameters used for compression and chunking for better results. We hope that this article also helps readers find those parameters.
It is worth noting that these techniques still have some limitations: they only work with contiguous slices (that is, with step 1 on every dimension), and on datasets with the same byte ordering as the host machine. Also, although results are good indeed, there may still be room for implementation improvement, but that will require extra code profiling and parameter adjustments.
Finally, as mentioned in the Blosc2 NDim post, if you need help in finding the best parameters for your use case, feel free to reach out to the Blosc team at contact (at) blosc.org.
Enjoy data!
Comments
Comments powered by Disqus